Background/Cell Mutations
Mutations can arise for a large number of reasons. The cell cycle is a very delicate process and there are several checkpoints that ensure, for example, that the DNA has been replicated correctly, prior to entry into the G2 phase of the cell cycle, or that the spindle fibres are correctly attached to the chromosomes prior to the initiation of anaphase (I/II).
Types of Mutation in DNA Replication
There are two main types of mutations: chromosomal and point.
Chromosomal Mutations
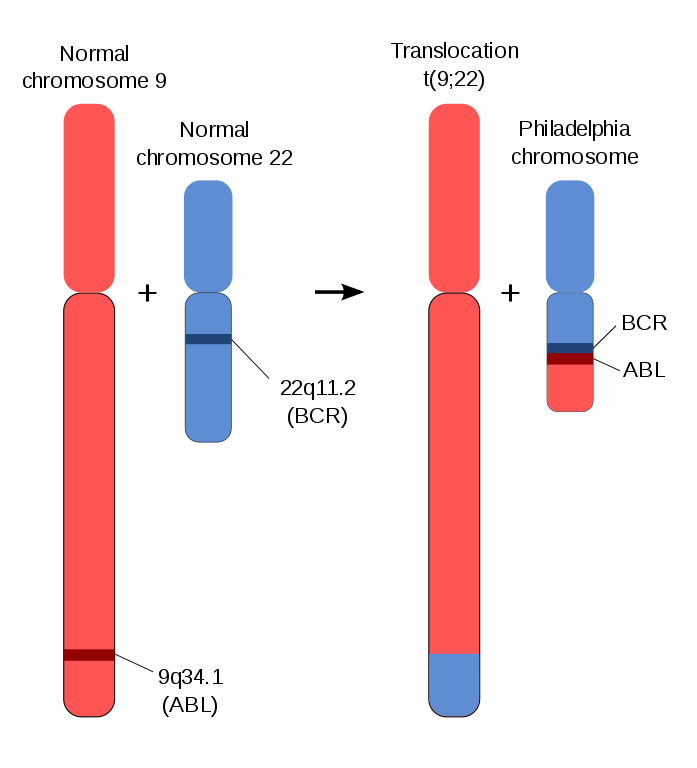
Figure 1: Chromosomal mutation [1].
In chromosomal mutations, individual components of a chromosome will be shifted from one location to another, e.g: on another chromosome, or lost all together. For example in Figure 1, a DNA segment from chromosome 1 translocates to chromosome 2, such the genetic material is swapped between the two chromosomes. This is different from the swapping of genetic material in meiosis as the exchange of genetic material occurs between different (non-homologous) chromosomes.
Point Mutations
To understand DNA point mutations we will consider one of the two strands of DNA. The most important elements of the DNA strand are the bases (A, T, C and G). As such, DNA strands are often described in terms of these letters only. It is possible to describe only one of the two DNA strands because the other strand can be determined by the basic rules of DNA structure: A pairs with T and C pairs with G.
So if one of the DNA strands had the letters …ATGCGAT… the other strand would have the letters …TACGCTA…
In a point mutation the actual DNA code changes. This may, or may not, have any effect; it will ultimately depend on the location of the base-pair change and the type of change. This will be explained shortly.
There are several types of point mutations:
- Duplications
- if the segment …ATGCGAT… underwent a duplication, it would read …ATGCGATATGCGAT…
- Deletions
- in a deletion, one (or more) bases are permanently removed. The segment …ATGCGAT… could read as follows after a single deletion: …ATGCGT…
- Insertions
- in an insertion, a new base may appear in the DNA such that …ATGCGAT… now reads …ATGCGTAT…
- Inversions
- a segment of DNA may flip such that …ATGCGAT… now reads …TAGCGTA…
- Substitutions
- in a substitution, one base may be replaced by another: …ATGCGAT… → …ATGCGGT…
These mutations may be spontaneous (occur without any outside influence) or induced. Spontaneous errors arise due to errors that arise during DNA replication (e.g: due to errors caused by the enzyme DNA polymerase). Often, spontaneous mutations are repaired, however they may persist. Induced mutations are those which are caused by an external stimulus. Examples of mutagens include radiation (e.g: UV light) and chemicals such as nitrous acid or benzopyrene.
Effects of Mutations
The DNA code is read in blocks of three: e.g: ATCGTG would be read: ATC then GTG. Each three letter block can encode a single amino acid. Long chains of triplet-bases will form long chains of amino acids, aka. proteins.
With 4 bases, there are a total of 43 (= 64) triplet combinations. However, there are only 20 biological proteins. As such, there are an excess number of triplet bases. The DNA code is thus called degenerate. This is not a concern, however, as two different triplet bases could encode the same protein: for example, the AGT and AGC both encode the protein serine. As a consequence of this, certain mutations (especially substitution mutations) may have no overall effect on the structure of a protein. (e.g: if the base sequence …AGT… mutated to give AGC, there would be no noticeable effect).
Other mutations may change a single amino acid, but have no noticeable effect on the overall structure or function of the protein because the new amino acid has similar chemical and physical properties to the original amino acid.
Other mutations may cause a noticeable biological effect. This would normally arise when the encoded protein takes on a different structure due to a change in the underlying amino acid properties.
It is also possible for a mutation to have no effect despite changing the chemical and physical properties of the amino acid. This would occur in non-coding regions of DNA (i.e: regions of DNA code that serve no apparent biological function). Mutations in these regions almost always go un-noticed.
Tumour Suppressor Genes and Proto-Oncogenes
Mutations of the following two genes are involved in the development of cancer: proto-oncogenes and tumour suppressor genes.
Proto-oncogenes encode for biological proteins that regulate cell division; they can mutate to form oncogenes which themselves promote tumour development. Only 0.1-0.2% of human genes are capable of becoming oncogenes [2]. If proto-oncogenes fail, tumour suppressor genes should activate and halt tumour development. Mutations of tumour suppressor genes may permit cancer development, although, normally, both alleles (copies) of a tumour suppressor gene must be mutated, as the tumour suppressor genes function recessively [3],[4]. In other words, if one tumour suppressor gene is functional, a mutation will not persist.
The immune system is the next barrier to tumour development. Mutations cause cells to produce proteins that the body recognises as foreign. The immune system normally attacks and eliminates these cells. For a cell to become cancerous it must be able to avoid detection and destruction by the immune system.
References
- Image taken from: https://en.wikipedia.org/wiki/Philadelphia_chromosome (accessed: 19/06/2016).
- Weinberg RA. Oncogenes and tumor suppressor genes. CA Cancer J Clin 1994;44:160–70. DOI:10.3322/canjclin.44.3.160.
- Weinberg R. Tumor suppressor genes. Science (80- ) 1991;254:1138–46. DOI:10.1126/science.1659741.
- Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3.