Clatterbridge/TOPAS
This simulation is a model of the 62.5 MeV ocular proton beam at the Clatterbridge Cancer Centre built using TOPAS, a wrap around of the Geant4 code specifically developed for proton therapy applications. This model was redeveloped in TOPAS to incorporate geometry updates and general progress made from the previous Geant4 model.
Parameter system
TOPAS uses a parameter system and employs a modular framework to assemble and execute a simulation. Lines of code do not need to be in a particular sequence as TOPAS does not enforce strict order dependence. Parameter files must be written and read in plain text format and their functions are specified given the following command structure:
Parameter_Type : Parameter_Name = Parameter_Value
Where the Parameter_Type is the declared object type: string (s), integer (i), boolean (b), unitless double (u), dimensioned double (d) or a type of vector i.e. integer vector (iv). The Parameter_Name is a sequence of a prefix related to functionality (i.e. Ge: Geometry), the object name and property. These prefixes enable different things to be controlled with a single line command, such as the physics (Ph), scoring (Sc) particle source (So), material (Ma) and graphics (Gr) etc.
File chain structure
As more complicated applications require hundreds of parameter lines, it is logical to split up the code into several parameter files. A file will inherit and execute all lines from other files which are called by:
includeFile = anotherParameterFile.txt
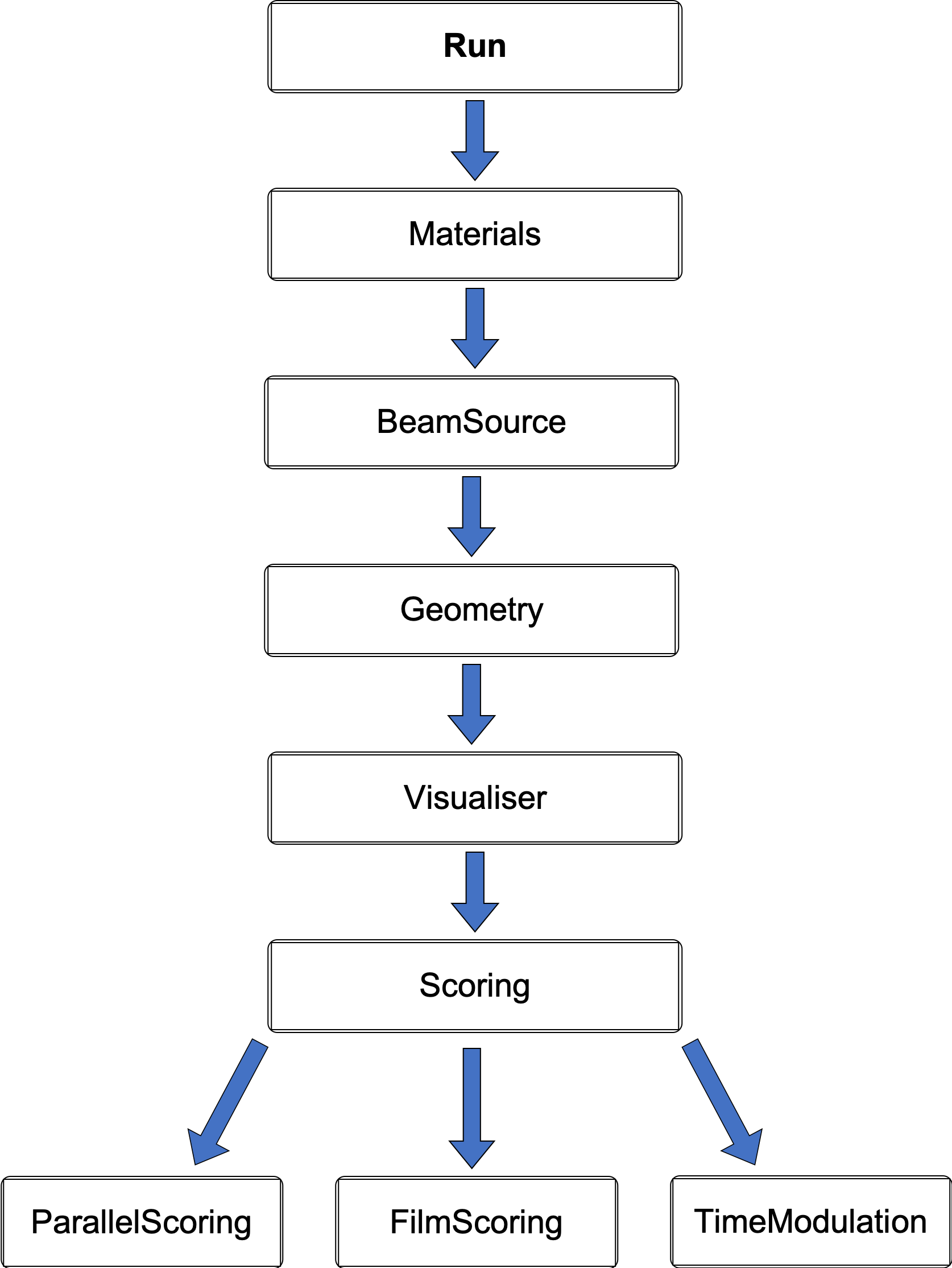
It can be useful to also call another file from within the secondary parameter file where it then becomes a parameter file chain. This allows the application to be organised and managed as essentially separate modules: a supportive feature for collaborative work or developing different aspects of a simulation model. Parameter file chains can span horizontally as well as vertically, however the hierarchy structure must be considered as the order is important in this case. Parameter lines higher up in the chain will take precedence over the same commands further down. Moreover, to handle all the different parameters, TOPAS does not allow settings belonging to the same object to be changed in a separate chain. Two different files are not able to modify the same parameter. Similarly, all components which are given the same geometry or the same scorer must be contained in the same vertical chain. Fundamentally, the main or default parameters should be defined in the initial file and then everything else that utilises these conditions should be written into adjoining files. A mostly linear hierarchical structure was adopted with interchangeable files for the scoring, this is illustrated in the diagram below.
Geometry
In general, TOPAS geometry can be described in different ways: essentially the dimensions and position must be specified with respect to the centre of their parent volume. The component name is included in each line with the stated type (shape), parent volume and material. Translational and rotational X, Y, Z parameters are assigned for positioning of the component within its mother in x, y and z. The orientation however will depend on the type of component and additional parameters may be required.
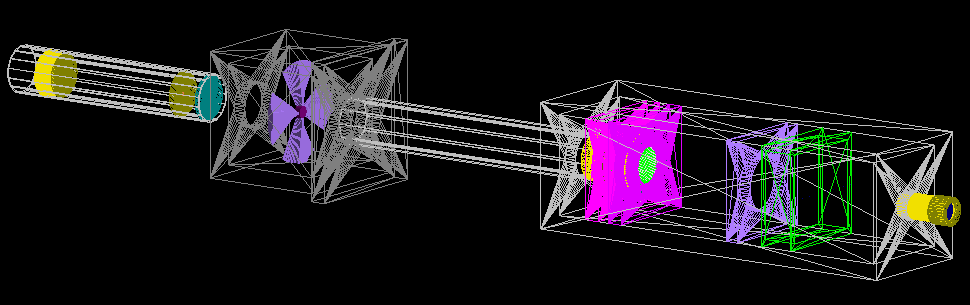
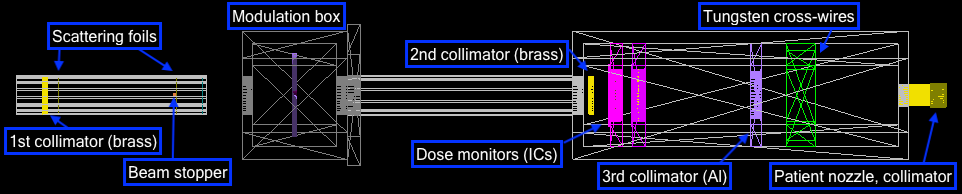
Treatment beamline
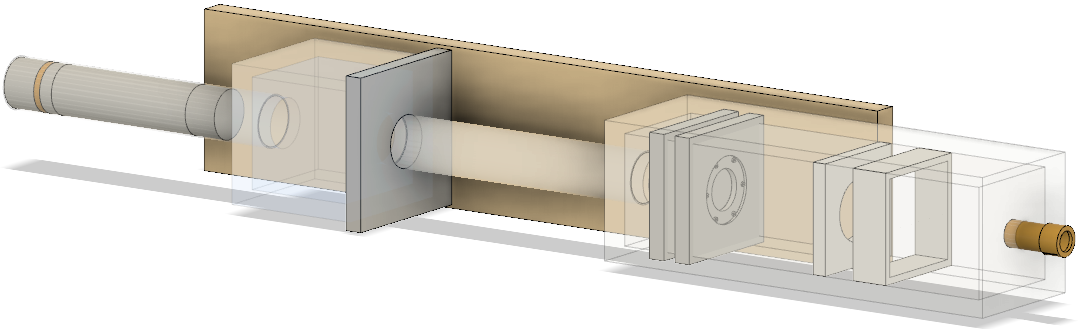
CAD model
The geometries were built in CAD according to their exact physical dimensions which are displayed in this complete schematic. Components are exported as Stereolithography binary (STL) files which describe geometry only by their surfaces (a tessellation of triangular and quadrangular faces which approximate the volume). Although other file formats (i.e. GDML) may intrinsically include different properties such as materials or colour attributes, TOPAS does not interpret this information and each component (with their elements grouped by material) exists as an individual STL file. Furthermore, components may have their own respective axis within the world coordinate system retained during export and additional rotation or translation using parameter lines may be required. To avoid overlaps, a 0.05 mm tolerance was built betwee adjacents surface and components. The positions of every single element within the treatment line coordinate system are displayed in this listing.
Physics settings
Visualisation
Particle Source
Phase space inputs
BDSIM
Scoring
Volume scorers
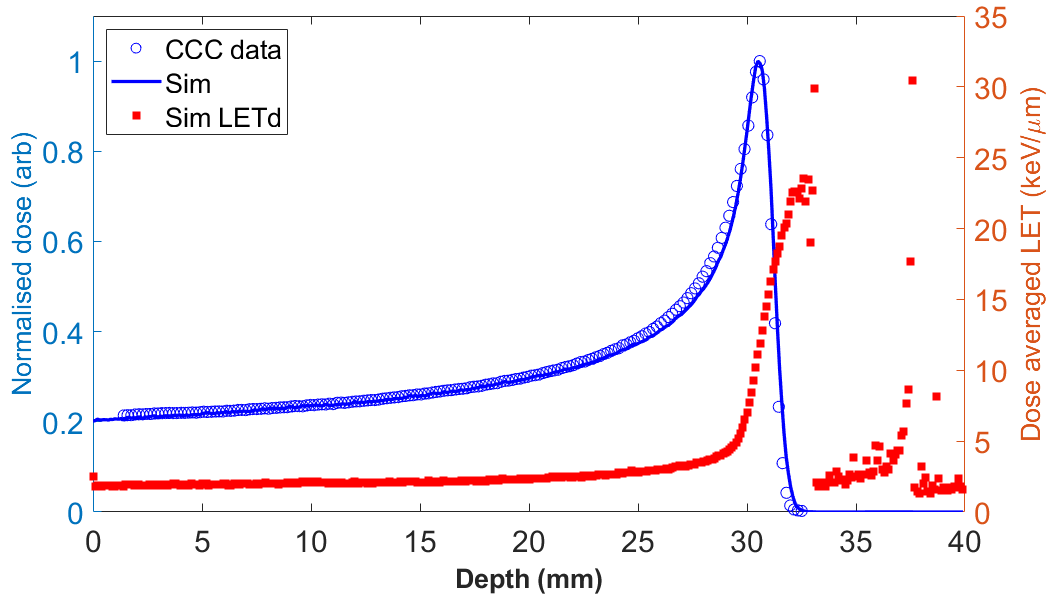
Dose deposition
Linear energy transfer
Surface scorers
Transverse beam profiles
Running the simulation
TOPAS
The simulation has been updated for compatibility with TOPAS v3.6.1 and can be cloned from this Github repository.
Output
Analysis
Dose profiles